First revision written in 2004.
Updated in 2013.
To calculate the Haar transform of an array of n samples:
- Treat the array as n/2 pairs called (a, b)
- Calculate (a + b) / sqrt(2) for each pair, these values will be the first half of the output array.
- Calculate (a - b) / sqrt(2) for each pair, these values will be the second half.
- Repeat the process on the first half of the array.
(the array length should be a power of two)
Example
Our input array will be these eight elements:
[ 1.000 2.000 3.000 1.000 2.000 3.000 4.000 0.000 ]
After calculating the sums and differences:
[ 2.121 2.828 3.536 2.828 -0.707 1.414 -0.707 2.828 ]
Recursing on the first half:
[ 3.500 4.500 -0.500 0.500 -0.707 1.414 -0.707 2.828 ]
Recursing again gives us the output array:
[ 5.657 -0.707 -0.500 0.500 -0.707 1.414 -0.707 2.828 ]
Haar basis functions
Taking the inverse Haar transform of one coefficient at a time shows the Haar basis functions. e.g.:
inverse([1 0 0 0 0 0 0 0]) = [ 0.011 0.011 0.011 0.011 0.011 0.011 0.011 0.011 ] inverse([0 1 0 0 0 0 0 0]) = [ 0.011 0.011 0.011 0.011 -0.011 -0.011 -0.011 -0.011 ] inverse([0 0 1 0 0 0 0 0]) = [ 0.050 0.050 -0.050 -0.050 0.000 0.000 0.000 0.000 ] inverse([0 0 0 1 0 0 0 0]) = [ 0.000 0.000 0.000 0.000 0.050 0.050 -0.050 -0.050 ] inverse([0 0 0 0 1 0 0 0]) = [ 0.224 -0.224 0.000 0.000 0.000 0.000 0.000 0.000 ] inverse([0 0 0 0 0 1 0 0]) = [ 0.000 0.000 0.224 -0.224 0.000 0.000 0.000 0.000 ] inverse([0 0 0 0 0 0 1 0]) = [ 0.000 0.000 0.000 0.000 0.224 -0.224 0.000 0.000 ] inverse([0 0 0 0 0 0 0 1]) = [ 0.000 0.000 0.000 0.000 0.000 0.000 0.224 -0.224 ]
Or, visually:
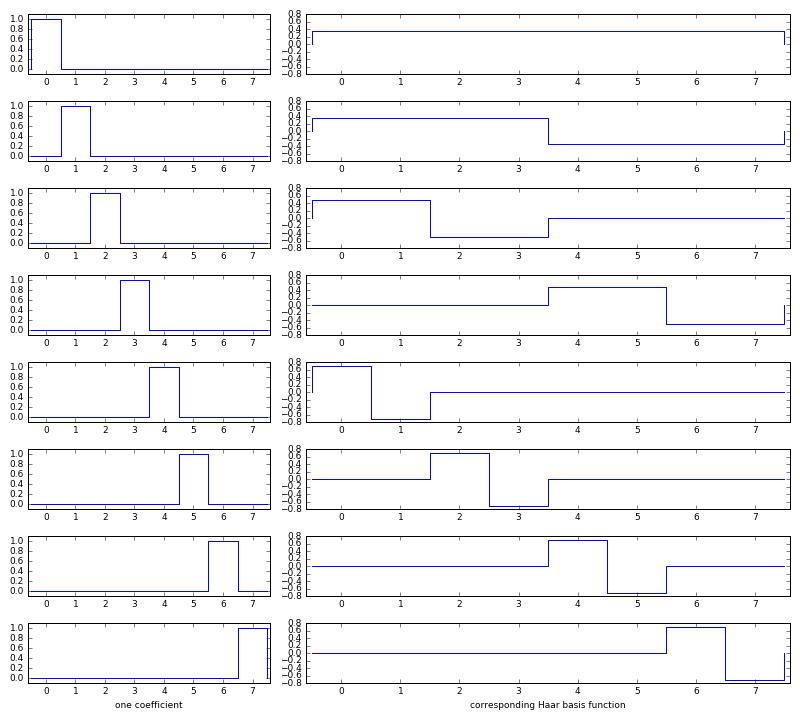
(source code: haar-basis.py)
The Haar basis functions are orthonormal. If we look at each basis function as a vector in eight-dimensional space:
- Each one is a normal vector. (i.e. Its length is one unit.)
- All of them are orthogonal (perpendicular to each other in eight-dimensional space.) (i.e. The dot product of any two basis functions is zero.)
The output of the Haar transform will have the same energy (same sum of squares) as the input.
2D Transform
Given a two-dimensional array of values, we can perform a 2D Haar transform by first performing a 1D Haar transform on each row:
→ → → →
And then on each column:
↓ ↓ ↓ ↓
Image Compression
The Haar transform can be used in lossy image compression:

→ 
1. Original image 2. Haar coefficients 
→ 
3. Fewer Haar coefficients 4. Reconstructed image
(source code: haar-lossy.py)
The blocky artifacts are interesting because they look so different from the JPEG artifacts I'm used to. In the above example, I threw away 95% of the coefficients, so that only the strongest (in terms of absolute value) remained.
This approach doesn't work so well with images that have strong noise:

→ 
before after
References
Image Compression Using the Haar Wavelet Transform
Also big thanks to Gabriel Peyré for answering my questions.